Understanding How Flu Viruses Change
Flu viruses have the capacity to change both slowly, through small genetic changes that are passed down to daughter generations, and quickly, through a process called “reassortment” that mixes larger genetic segments from several viral strains to create a new virus. Both processes are important to influenza’s success as a disease-causing organism during flu seasons and in pandemics.
This page provides an overview of what scientists have learned about the basic mechanisms behind the influenza virus’s ability to change. Also, this section will help you sort through the major terms used in the discussion, from genetic shift and genetic drift to triple reassortment.
Not all changes are created equal (antigenic drift)
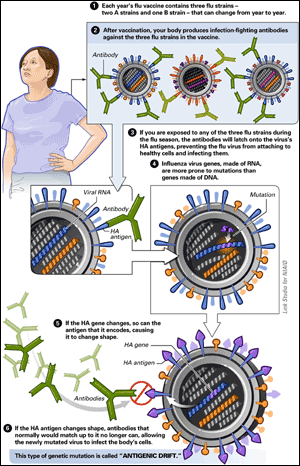
To understand the major genetic changes that lead to a pandemic, it helps to first understand the minor genetic changes that produce new flu strains in each flu season. Like all living things, influenza makes small errors—mutations—when it copies its genetic code during reproduction. But influenza lacks the ability to repair those errors, because it is an RNA virus; RNA, unlike DNA, lacks a self-correcting mechanism. As a result, influenza is not genetically stable. Every generation is slightly different, and those differences accumulate as time passes.
That slow deviation is called
antigenic drift, and it is the reason why it is necessary to reformulate flu vaccines every year (see figure below). Every flu season, the genetic make-up of the dominant strains from the prior year will have drifted, changing the surface structure of those strains just enough to diminish, or even destroy, the effectiveness of the previous year’s vaccine. Each winter, health authorities must make an educated guess which strains are likely to dominate in the next flu season. It takes six more months to develop and manufacture vaccines for chosen strains. But in some years, genetic drift during just those six months can render the newly formulated vaccine ineffective, leaving populations more vulnerable to the newly evolved virus.
ANTIGENIC DRIFT:
 | View larger image |
| View larger image |
“Antigenic” refers to proteins on the virus’s surface that allow the virus to infect its host and that cause the host’s immune system to react. Antigenic drift can occur in all three types of influenza: A, B, and C.
An eternal exchange
Antigenic drift is slow and moderately predictable: Researchers understand the rate at which flu changes, though they cannot predict how different the final product will be. But flu can also change rapidly, due not to mutation, but to what is called a “genetic exchange event,” in which flu viruses in close proximity swap entire sections of their genetic code. “Genetic-exchange events” are particularly important in the making of new pandemic strains.
It may help to think of viruses not as single, fixed species but as a dynamic population of numerous variants that undergo selective pressure. When different populations can coexist in multiple susceptible mammalian hosts (humans, pigs, cats), there is an increased likelihood of mixing of genetic material in recombination events. Scientists are only beginning to understand the molecular mechanisms underlying the selection process, including the change in the efficiency of human-to-human transmission, which is a major factor in turning an epidemic strain into a pandemic one.
1
The significance of antigenic shift
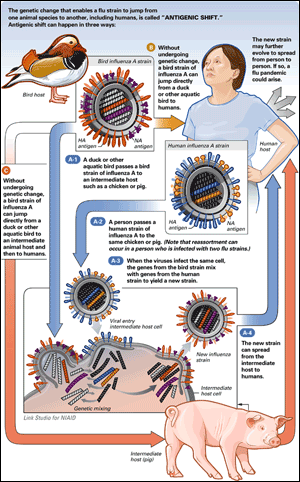
Rapid change in circulating flu viruses is known as antigenic shift. Shift can happen in several ways: when a flu strain jumps to a species which it has never before infected, or when the process of viral reproduction creates a novel strain. In either case, the human immune system perceives the strain as unfamiliar. And that lack of immunologic experience is what causes the much larger amounts of illness that are created by a shifted strain.
Antigenic shift can occur in three different ways, as explained and illustrated in the figure below:
ANTIGENIC SHIFT:
 | View larger image |
| View larger image |
Antigenic shift occurred in 1997 when the H5N1 avian flu virus appeared for the first time in humans, one example of what happens when a flu strain jumps from one animal species to another, or from an animal species to a human host. (See also
Crossing the Species Barrier.)
Antigenic shift can also occur when at least two influenza A strains infect animals simultaneously and combine to form a new virus that contains a mixture of the genes of the two original strains. (Scientists call that antigenic shift via
reassortment.) This can also happen in people infected with two strains at the same time, although birds and
swine are considered the major “mixing vessels.”
New influenza types emerge from such unpredictable recombination—reassortment—events between human and mammalian or avian viruses. These events occur at irregular intervals.
2 If the new virus causes illness in people and can be transmitted easily from person to person, then the virus has the potential to cause an epidemic or pandemic.
3
How reassortment creates an entirely new virus
Antigenic shift from reassortment can produce major changes in the influenza virus and represents a significant way for viruses to evolve and create a new pandemic strain.
Influenza’s genome is made up of eight loosely linked segments, each of which harbors at least one important gene. Those genes direct the expression of influenza’s major viral proteins, including hemagglutinin and neuraminadase, the antigenic proteins on the viral surface that that foster infection and stimulate immune reaction.
When flu viruses infect cells to reproduce, they use the cell’s molecular machinery to manufacture individual virus components including more viral RNA and proteins, and package them into new viruses that bud off of the cell. In the process of reproduction, the linkages between the segments of the flu genome break apart, leaving the segments free.
If the virus happens to have infected a cell that is simultaneously being infected by a flu virus of another strain, then the possibility exists that some of the genetic segments from those strains will be swapped in the process of reproduction. For instance, if a human H3N2 flu virus and a bird H7N3 flu virus infect a person, reassortment can intermingle genes from both viruses during replication.
4 Because more than one virus is being produced in the infected cell, new viral copies could have a hemagglutinin (H7) from the avian virus and neuraminidase (H2) gene from the human virus (see
Figure: Antigenic shift), making a reassortant flu virus, H7N2, that is different from the two original viruses.
The significance of these events is that they create influenza A viruses with different properties from the original infecting strains. The reassortant virus could, for instance, possess an avian hemagglutinin (H) protein against which humans have little or no immunity plus human influenza genes that are more likely to cause sustained human-to-human transmission. The net result could be a new strain that has pandemic potential.
Other types of reassortment events can shuffle individual viral genes. Because flu’s genome has eight segments, the potential number of different viral progeny can be as great as 256 types (28). Much of this information comes from the ability to sequence viral genetic material and compare sequences between different viruses.
Pandemics from reassorted viruses
Reassortments among subtypes from avian and human viruses were important in producing new strains that caused the 1957 (H2N2 subtype) and 1968 (H3N2 subtype) human influenza pandemics.
Multiple reassortment events are also possible and have prompted concerns about the creation of new viruses. Researchers had thought that only the H and N gene were switched during reassortment. Recent analyses of influenza genetic material suggest that multiple genes can be recombined. Several investigators have found influenza viruses that arose from triple reassortment events among swine.
5
Genetic sequence analysis of many viral subtypes suggest that the current pandemic swine influenza virus arose from a reassortment event between a triple reassortant influenza virus containing human, swine, and avian influenza genes and a Eurasian swine influenza virus.
6
Of the eight genes in the influenza 2009 A (H1N1) strain causing the current human pandemic:
| • |
PA, PB2 are swine genes that originated in birds (a triple reassortant strain) |
| • |
PB1 is a swine gene that passed from birds to humans then swine (a triple reassortant) |
| • |
HA, NP, NS are swine genes that originated in birds (classical swine virus/triple reassortant). |
| • |
NA, M are Eurasian swine genes that originated in birds (Eurasian swine influenza virus).7 |
Sources
-
Mark S. Klempner, Daniel S. Shapiro, “Crossing the Species Barrier — One Small Step to Man, One Giant Leap to Mankind,” New England Journal of Medicine 350:12 (March 18, 2004): 1171-1172 ↑
-
K. Stöhr, “Influenza,” Control of Communicable Diseases Manual, ed. David L. Heymann (Washington, DC: American Public Health Association, 2004) ↑
-
Centers for Disease Control and Prevention, Transmission of Influenza A Viruses Between Animals and People, Oct. 17, 2005. ↑
-
Ibid. ↑
-
Jon Cohen, “New Details on Virus's Promiscuous Past,” Science 324 (May 29, 2009): 1127. ↑
-
Rebecca J. Garten, C. Todd Davis, Colin A. Russell, et. al., “Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans,” Science 325 (July 10, 2009): 197-201. ↑
-
Ibid. ↑